
Description
DeepChek®-HCV Software (CE-IVD) is a flexible and customizable downstream analysis software able to handle sequencing data and intended to be used for performing Viral Hepatitis C (HCV) genomic data analyses and clinical interpretations.
Compatible with Electrophoresis Capillary and Next Generation Sequencing (NGS) platforms (like MiSeq, MiniSeq, iSeq-100, 454-Junior, 454-FLX…), DeepChek®-HCV stores, organizes sequencing data in a dedicated database format and performs bio-informatics analyses and clinical interpretations of 5’UTR, CORE, NS3, NS5A or NS5B sequencing data. It is able to generate reports to be used either for research or for routine use.
- Supported formats: AB1, FASTA and PLAIN-TEXT for Sanger, FASTA/FASTQ (including paired sequencing), BAM/SAM for NGS covering Targeted Genes or also compatible with Whole Genome data.
- Available analyses: subtyping/high-resolution subtyping, amino-acid mutations/nucleotide changes determination and validation, NGS coverage validation
- Available interpretations: drug resistance assessment (for NS3, NS5A and NS5B inhibitors) from several up-to-date guidelines (including but not restricted to geno2pheno…)
- Included services: updates/upgrades (quarterly releases), support, web-training, historical sequencing data import
- On-demand services: local training, integration with third-party instruments or systems, customization…
The DeepChek®-HCV software system is a secured web application which can be used through a Cloud access or locally, through pre-configured servers. It is made available with regular updates (new clinical databases, guidelines…) and quarterly upgrades (new features, modules, applications…) and can be fully integrated within the IT network of each laboratory (integration with the sequencing platform, with the Laboratory Information System – LIS, with the Hospital Information System – HIS…).
For “In-Vitro Diagnostic use (IVD)” where indicated : CE marking only valid for EEA and territories recognizing it. Otherwise “For Research Use Only” (RUO): not for use in diagnostic procedures, no claim or representation is intended to provide information for the diagnosis, prevention, or treatment of disease.
Limitations
1. DeepChek® Software is a downstream analysis software program (“Program“) which enables virologists to input pre-formatted sequences from genetic analyzers (Sanger or Next Generation Sequencing) using either CE-IVD genotyping assays (“IVD information”) or for Research Use Only (“RUO information”) genotyping assays (“PCR amplification”) in order to obtain viral or bacterial sequence analysis and viral or bacterial drug resistance interpretations or other interpretations to adapt accordingly patient’s anti-viral or anti-bacterial drugs (“Analyses“).
2. For In Vitro Diagnostic Use only with IVD information or with combination of IVD information and RUO information. For research use only with RUO information alone.
3. Responses to anti-viral and anti-bacterial treatment are complex and affected by a number of factors not taken into account by the Program.
4. The selection of drugs for the treatment of viral or bacterial infection is the responsibility of the physician in consultation with the patient and reliance should not be placed on the Analyses only for such purposes.
5. The Analyses are not intended to replace professional medical care and attention by a qualified medical practitioner and consequently ABL does not accept any responsibility for the selection of drugs and the patient’s response to treatment.
6. As the accuracy of the results highly depends on the sequencing technology and on its related technical recommendations (Ex.: In case of ‘low’ viral load input, the users should be aware of risks of resampling errors), ABL cannot take any responsibility on the reliability of the results if the recommendations of the suppliers are not strictly followed.
7. ABL does not accept any responsibility for the accuracy of the data entered by the user or the consequences of any inaccuracies in those data.
Overview of the pipeline
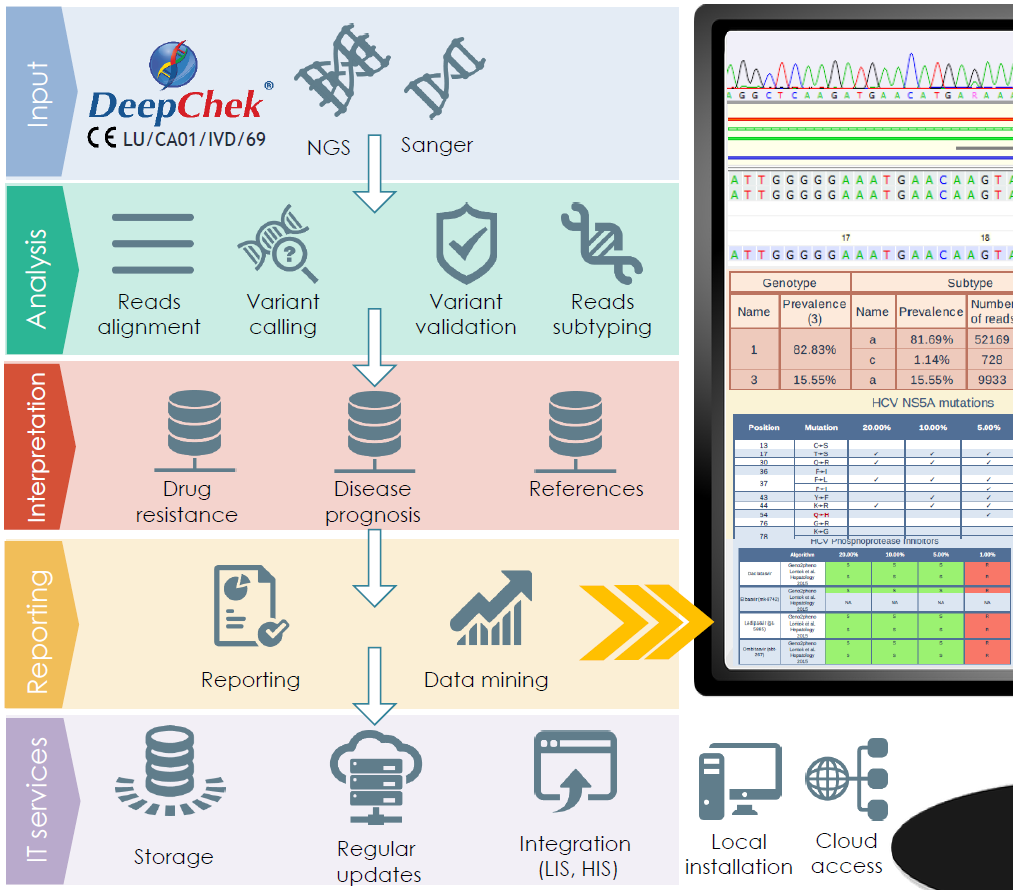
Characteristics and performances
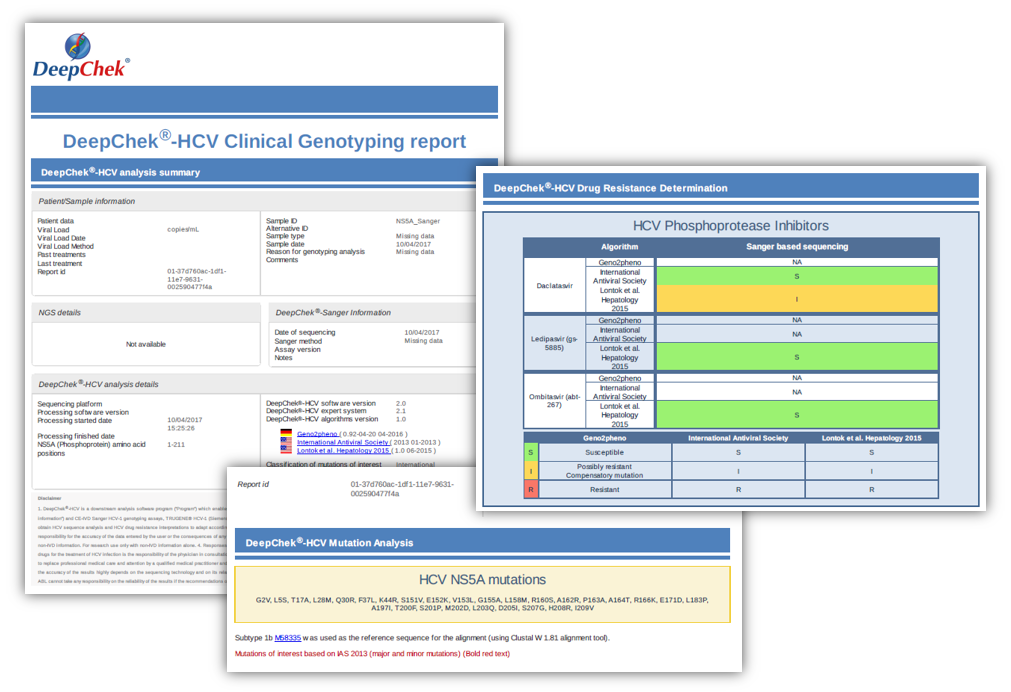
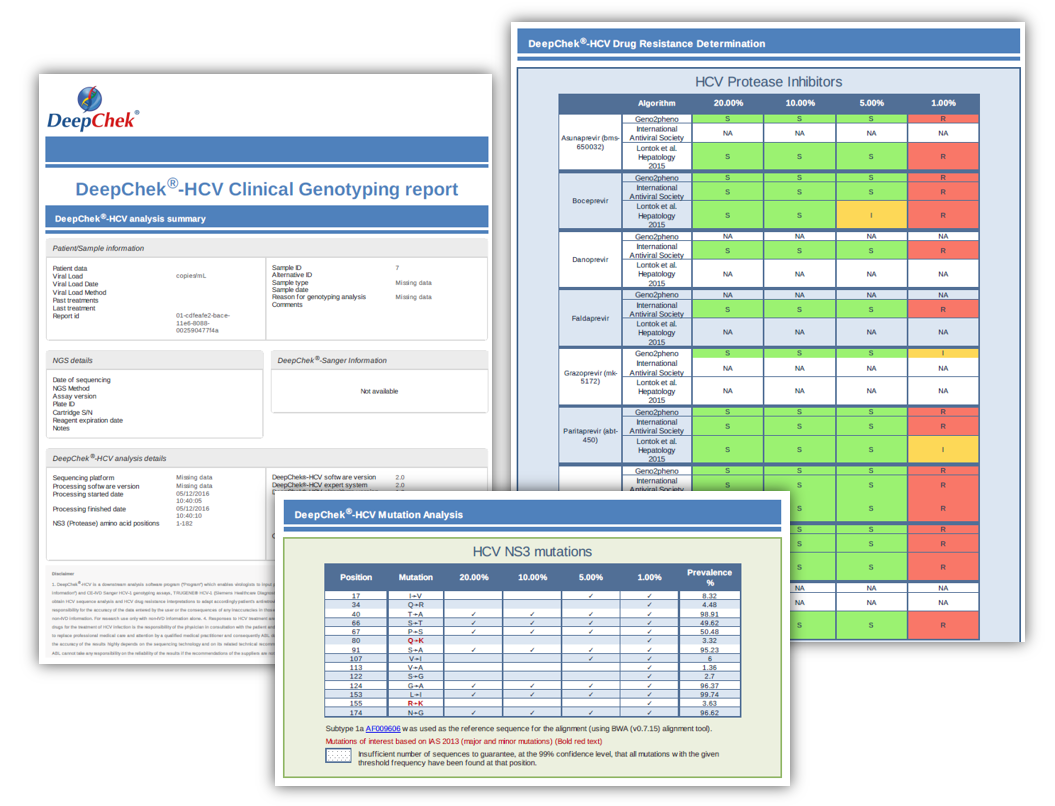
Examples of reports
Ordering information
Downloads
General documentation
-
Quick start guide
-
Implementation options

