Description
The DeepChek® Assay Whole Genome SARS-CoV-2 Genotyping is a RT-PCR Assay (nucleic acid technique (NAT)) followed by Next Generation Sequencing (NGS) intended to aid clinical applications and for use on previously diagnosed COVID-19 patients.
Application (RUO) — REF 159D48
For Research Use Only (RUO). Not for use in diagnostic procedures. No claim or representation is intended to provide information for the diagnosis, prevention, or treatment of disease.
The DeepChek Assay Whole Genome SARS-CoV-2 Genotyping (RUO) is a reverse transcriptase (RT) polymerase chain reaction (PCR) test (nucleic acid technique (NAT)) intended to screen the emergence of SARS-CoV-2 genome mutations using extracted RNA from patients already diagnosed PCR positive to SARS-CoV-2.
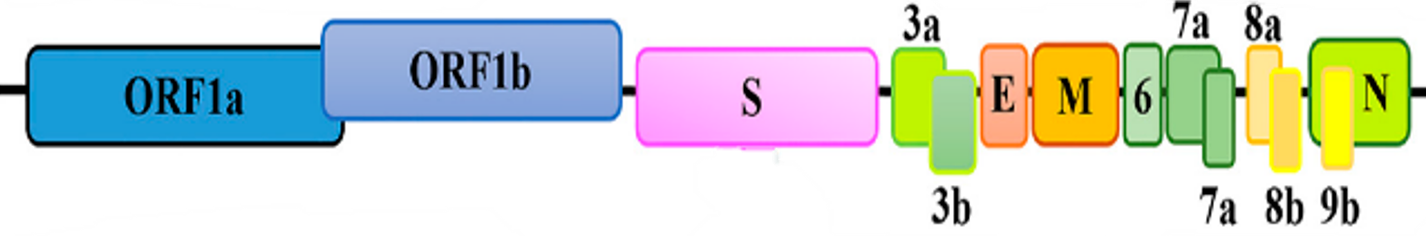
The test is amplifying the whole genome of the SARS-CoV-2, including regions which harbor mutations described as sufficient, when present, to characterize some variants.
Results are used to build basic research knowledge and evidence on the emergence of circulating SARS-CoV-2 variants.
Results are used to better understand SARS-CoV-2 transmission and to monitor for the emergence of new variants, even in minor viral populations, through ongoing epidemiological surveillance and strategic testing; conducting outbreak investigation and contact tracing; and where appropriate, adjusting public health and social measures to reduce transmission of SARS-CoV-2.
The DeepChek Assay Whole Genome SARS-CoV-2 Genotyping (RUO) is intended for use by trained clinical laboratory personnel specifically instructed and trained in the techniques of RT-PCR and next generation sequencing (NGS) workflow.
Methodology
Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) and Next Generation Sequencing.
The Assay targets and amplify the whole genome (>99%) of SARS-CoV-2 virus, including regions that harbor mutations of interest described as sufficient in databases like Pangolin and Nextclade, including variants of concern (VOC).
Post PCR – Complementary information
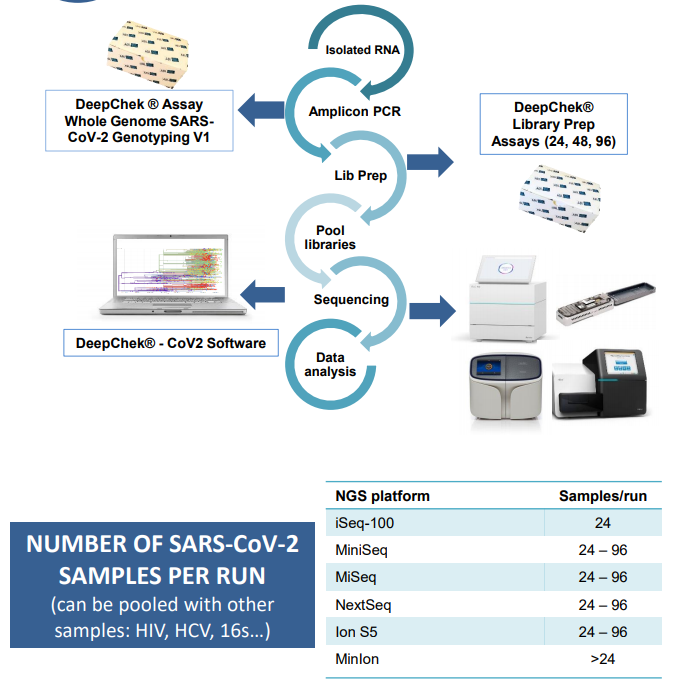
Next Generation Sequencing
The main volume of the product output is then used for next generation sequencing (NGS). The NGS workflow can be
different as the NGS technics and NGS analyzers vary. NGS sequencing instruments are general laboratory use devices.
Sequencing and library preparations reagents are general laboratory use products.
While the verification studies, we used the Illumina iSeq 100 Sequencing System (#20021532) and the following combination
of reagents: ABL DeepChek® NGS Library preparation (catalog #116BX, 24 or 48 or 96 tests), ABL DeepChek® Assay Adapters (catalog #124BX, 1-24, 1-48 or 1-96) and Illumina iSeq 100 Reagent (catalog # 20021533, 300 cycles).
Downstream NGS Analysis Software
The next generation sequencing raw data are uploaded in a specific downstream software tailored for SARS-CoV-2 analysis which is a standalone medical device (i.e. DeepChek CoV2 software (RUO) license and module (REF S-12-023 (CVL) and S-12-023 (CVM)). The software itself could be CE-IVD marked and shall refer to an updated expert list of mutations and variants per virus lineages with latest knowledge and analyses.
More information on the DeepCheck® SARS CoV-2 Whole Genome – Click here
Characteristics and performances
Workflow
Ordering information
Downloads
DoC
-
EC Declaration of Conformity