
Description
As treatment options for the management of Hepatitis B continue to grow, the clinician’s challenge to manage drug selection in the face of single and multi-drug resistance is increasing daily. Based on a dynamic global mutational database, SeqHepB has been designed to enable clinicians to manage their patients’ treatment regime on an informed and accurate basis. Genomic sequence analysis software within SeqHepB rapidly and accurately identifies those mutations and sub-mutations that lead to drug resistance and multi-drug resistance.
The SeqHepB system is a highly accurate and unique bio-informatics viral genomic sequencing software, which has a wide spectrum of applications, including for use by/for:
- Clinicians
- Laboratories
- Clinical Trials
- Drug Development
SeqHepB provides an on-line predicative tool, enabling clinicians to make an informed decision as to drug selection based on the mutational and resistance profile of the hepatitis B virus (HBV) in an individual patient at any given time.
SeqHepB is linked to a comprehensive in-vitro phenotypic database of major Hepatitis B drug resistant mutants and is built on the world’s most complete collection of patented HBV drug resistance mutations. It is underpinned by a consortium of experts in the management of hepatitis B, thus further supporting its own extensive clinical case studies and clinical utility.
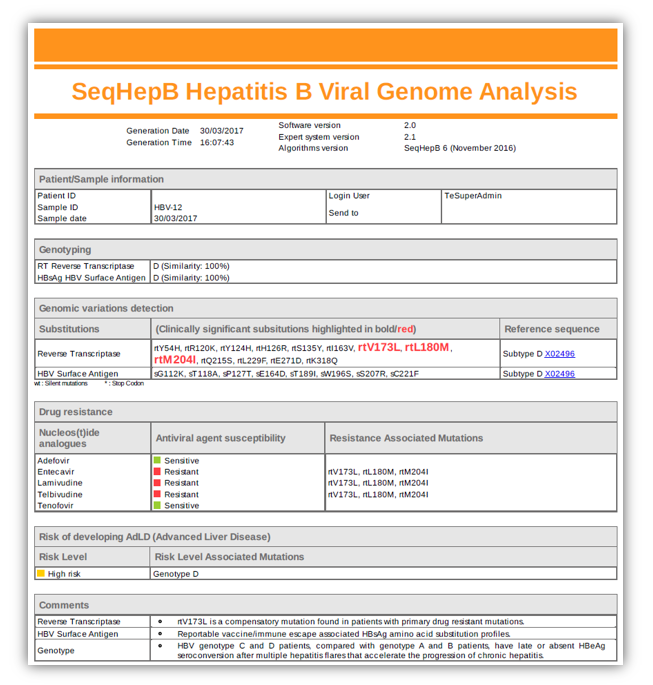
In practice, upon receipt of a patient blood sample, a licensed laboratory generates a genetic sequence which is then entered into the SeqHepB on-line system. The software generates a resistance profile based on the primary and secondary mutations found in the sequence.
As a dynamic HBV resistance profiling system, SeqHepB has been designed to keep up with a virus that is continually changing. It is this unrelenting evolution during antiviral therapy that is at the centre of cross-resistance and multi-drug resistance and enables SeqHepB to map the mutational “pathways” of the virus.
Access to SeqHepB
Characteristics and performances
Examples of reports
Ordering information
Downloads
General documentation
-
Quick start guide
-
Implementation options

