Description
The DeepChek® Assay-HCV NS5A Drug Resistance (RUO) is intended to be used for HCV drug resistance assessment. It provides drug susceptibility information for viral NS5A inhibitors. It combines target-specific PCR reagents with in vitro diagnostic software both compatible with either Sanger or Next Generation Sequencing platforms.
Assay should be used for patients with documented HCV genotype 1 to 6 (pan-genotypic assay).
For genotype 2 samples, in case of failure, please use the DeepChek® Assay NS5A (GT2) Drug Resistance V1 Assay(106A24) .
Methodology
DNA Sequencing • Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
Clinical Significance
EASL Recommendations on Treatment of Hepatitis C 2016: “Physicians who have easy access to a reliable test assessing HCV resistance to NS5A inhibitors (spanning amino acids 24 to 93) can use these results to guide their decisions, as specified in these recommendations. The test should be based on population sequencing (reporting RASs as “present” or “absent”) or deep sequencing with a cut-off of 15% (only RASs that are present in more than 15% of the sequences generated must be considered)”.
More information on the DeepChek® Assays – Click here
More information on the DeepChek® Software – Click here
Characteristics and performances
Features
Compatibility with Capillary Electrophoresis (SANGER) platforms
Any
Compatibility with Next Generation Sequencing platforms
Any
Intended use
RUO – ISO-9001 manufacturing
Types of samples
Serum, plasma, DBS
Format
24 samples/kit
Content
RT-PCR & Nested-PCR reagents (enzymes, master mixes, primers, dNTPs...), SANGER sequencing primers, protocols for SANGER and NGS sequencing, SANGER or NGS software analyses for 24 samples through a Cloud access
Sensitivity
1000 UI/mL for 400 µL plasma/serum. Protocols for low viral loads (>200 - 300 UI/mL) available
Specificity
Validated on all genotypes (pan-genotypic)
Reproducibility
>99%
Workflow
From sample to result in ~15 hours for SANGER and ~30 hours for NGS (depending on the platform)
Covered positions
NS5A: codons 1 to 222
Compatible extraction methods
Automatic (MagNA Pure Compact Nucleic Acid Isolation Kit I - Roche, Promega - Abbott), Manual (Manual extractions using QIAamp® Viral RNA - Qiagen)...
Data analysis and interpretation software
Included (DeepChek-HCV / CE-IVD)
Available analyses
Genotyping/Subtyping, amino-acid mutations detection, nucleotide changes detection, drug resistance, NGS run quality report, clinical genotyping report...
Drug resistance
Flexible, through up to 3 different up-to-date guidelines including geno2pheno...
Included services
Unlimited updates & upgrades of the software, support, training...
Options
Local servers, historical data import, integration with LIS and HIS, integration with sequencers, automation of the IT workflow, customization...
Examples of reports
For SANGER sequencing
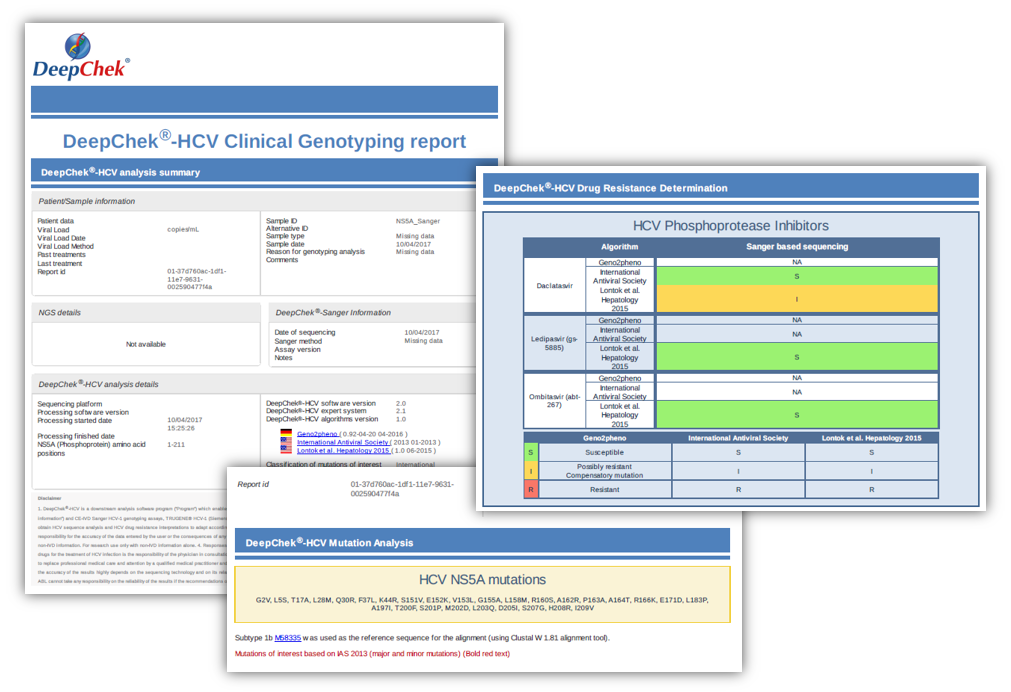
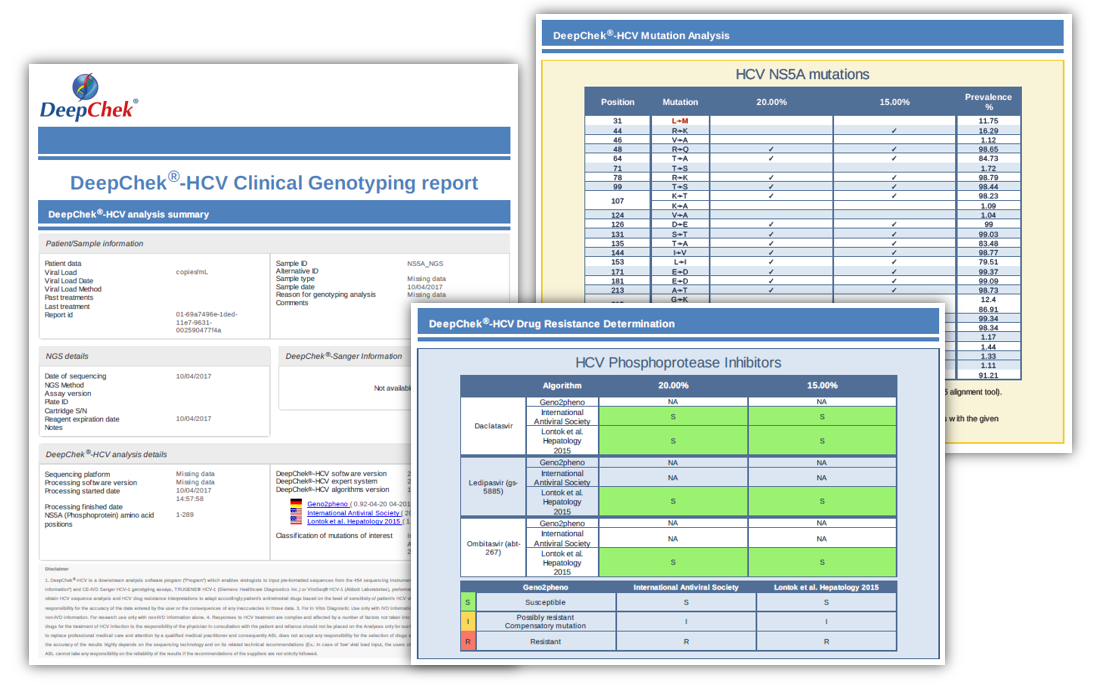
For NGS sequencing
Ordering information
Product
Reference
DeepChek® Assay-HCV NS5A Drug Resistance (RUO)
105A24
Downloads
General documentation
-
Protocol
-
Installation check list
-
Q&A
MSDS
-
International
Posters
-
Retreatment with Direct Active Antivirals of Genotype 1, 3 and 4 Chronic Hepatitis C Patients who Previously Failed an Anti-NS5A-Containing Regimen in Real World