
Description
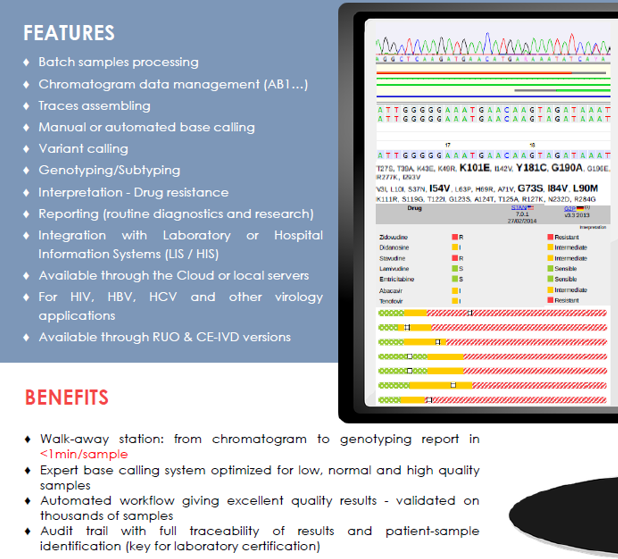
ViroScore®-HIV / ViroScore®-HIV Premium Software (S-09-014) is a downstream analysis software developed by ABL, able to handle SANGER sequencing data and intended to be used for performing genomic data analyses (genotyping, subtyping, amino-acid mutations detection, nucleotide changes characterization…) and clinical interpretations (drug resistance,…).
Compatible with Electrophoresis Capillary platforms, ViroScore® stores, organizes sequencing data in a dedicated database format and performs several types of analyses and interpretations to generate reports to be used either for research or for routine use.
- Supported formats: AB1, FASTA and PLAIN-TEXT
- Available analyses: subtyping of Pure or CRFs strains, amino-acid mutations/nucleotide changes determination and validation
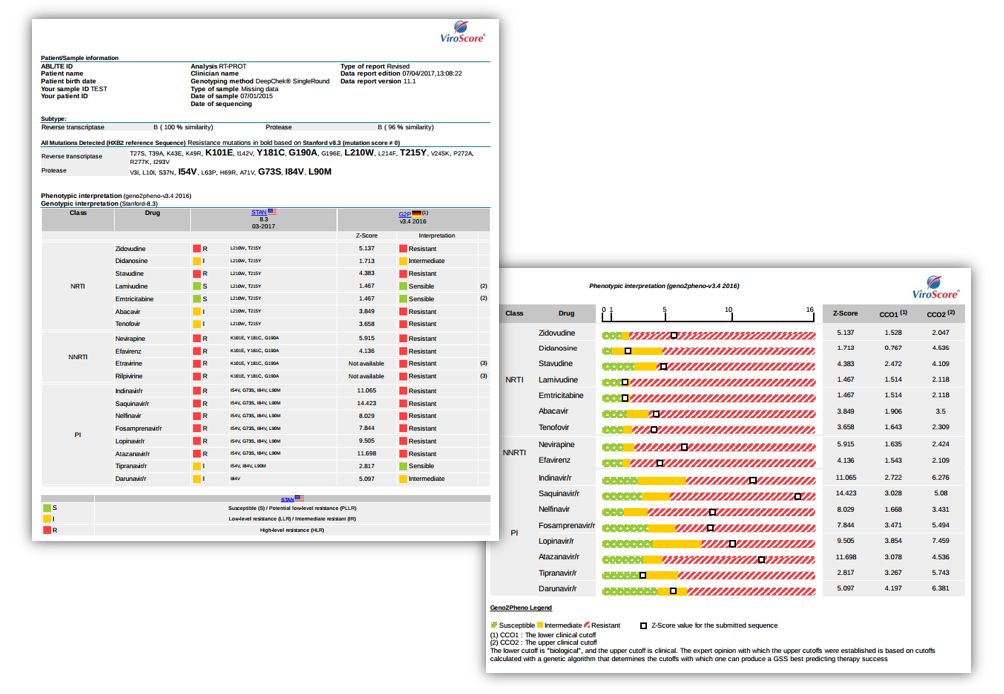
- Available interpretations: drug resistance assessment (for NRTIs, NNRTIs, IIs, PIs, EIs, FIs) from genotypic of virtual-phenotypic based guidelines (including but not restricted to HIVdb/Stanford, ANRS, geno2pheno, Rega…), tropism determination, Genotypic Sensitivity Scoring (GSS)…
- Included services: updates/upgrades (quarterly releases), support, web-training, historical sequencing data import
- On-demand services: local training, integration with third-party instruments or systems, customization…
The ViroScore® software system is a secured web application which can be used through a Cloud access or locally, through pre-configured servers. It is made available with regular updates (new clinical databases, guidelines…) and quarterly upgrades (new features, modules, applications…) and can be fully integrated within the IT network of each laboratory (integration with the sequencing platform, with the Laboratory Information System – LIS, with the Hospital Information System – HIS…).
The Premium version of ViroScore®-HIV is able to automate the entire bio-informatics workflow starting from electropherogram data in AB1 format to the clinical report. The entire analysis can be tackled in no more than a minute.
The ViroScore®-HIV application is CE-IVD (LU/CA01/IVD/70).
Overview of the pipeline
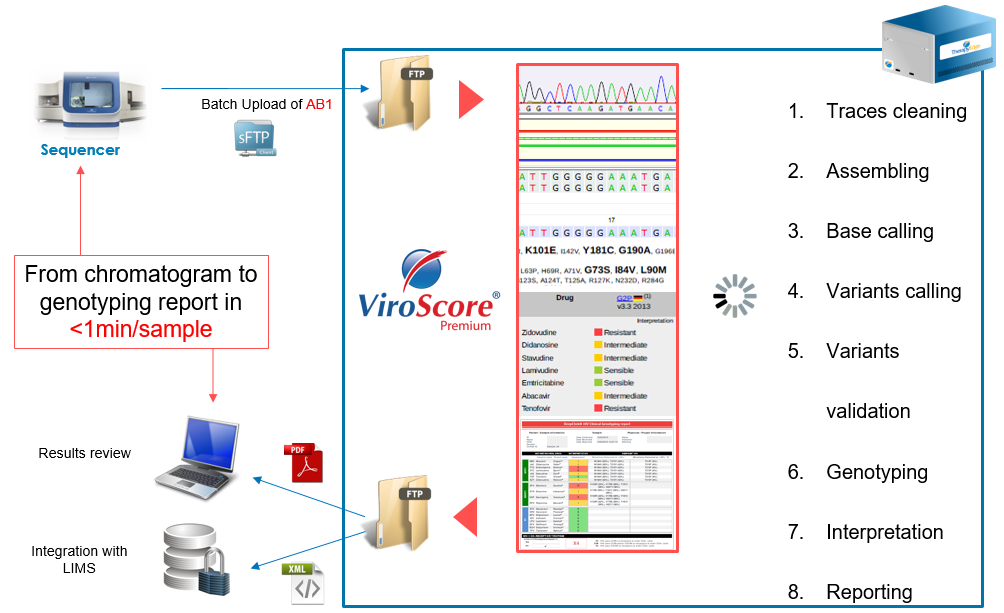
Characteristics and performances
Examples of reports
Ordering information
Downloads
General documentation
-
Brochure
-
Implementation options
Posters
-
Comparison of interpretive applications used for HIV-1 drug resistance determination
-
Comparison of HIV-1 Drug Resistance Interpretations Software
-
Software Dependent Differences in HIV-1 Drug Resistance Determination
-
Validation of New Informatics Systems for Routine HIV-1 Genotypic and Virtual Phenotypic Antiviral
-
Comparison of HIV-1 drug resistance profiles generated from novel software applications for routine patient care
-
Use of Innovative Information Systems Combining HIV-1 Genotypic and Phenotypic Drug Resistance Interpretations

